Solution: BLASTED
Answer: BIOMARKER
Written by Grant Yang
The puzzle consists of a long DNA sequence. The title and flavortext clue that this is actually a long BLASTn query sequence . Entering the entire sequence into the BLAST database yields various results with high similarity, although none of these cover the entire sequence entered. By analyzing the lengths and placements of each match, the long sequence can be divided into 9 different genes.
Each gene has a few insertions in it when compared to the database sequences, and the insertions appear as one or two letters. The flavortext clues that these are morse code, with one letter representing a “dot” and two letters representing a “dash”, and two spaces between insertions representing a break between letters.
The insertions for CEBPA:
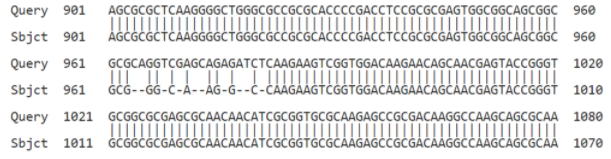
| HGNC Gene Symbol | Insertions (in lowercase) | Morse | Translation |
|---|---|---|---|
| CEBPA | …caGGtCgAgcAGaGatCt… | - ..- .-. | TUR |
| CLIC1 | …gcTgTTgc… | -. - | NT |
| IPO4 | …gCaCaGGaaCCtgCaaGtc… | ... - --- | STO |
| DNMT3A | …cGtGatCCaCtCg… | ..- ... | US |
| NFATC1 | …aGccGGcaGtGCatCtGc… | .- -. -.. | AND |
| ARRB2 | …gtCCgCatGc… | - .-. | TR |
| ALKBH1 | …tCggCCaaTa… | .- -. | AN |
| UBE2A | …cGcTaTTcTcgTaCa… | ... .-.. | SL |
| CNR2 | …cCAgtAAtcGAc… | .- - . | ATE |
The morse code says: TURN T’S TO U’S AND TRANSLATE. Changing all the T’s in the insertions into U’s and combining them yields:
CAUGGCAAUUGCGGCGAAAAUGAAUCCUAUAUGACCCAUAUUCGUGAUCUGGAAACCACCGAACGUUCC
Translating this into amino acids and taking the one letter symbol yields: HGNC GENE SYM THIRD LETTERS. This tells you to look at the third letters of the official HGNC gene symbols for the genes in the query sequence, yielding the answer: BIOMARKER
Author’s Notes
I wrote this puzzle as an homage to my bio classes and having to sit through minute-long query searches on the NCBI website. I wanted the solve experience to echo the process of geneticists obtaining a long DNA sequence and deciphering where genes start and end, as well as identifying indels through BLAST. Getting BLAST to give consistent mismatch reports proved a little difficult, requiring at times to select genes with CG-rich regions to insert A and T bases, and vice versa.
The partial answer TURN T’S TO U’S AND TRANSLATE is a little redundant if you know how to translate DNA to amino acid, but it helps to confirm the insertions and makes sure teams don’t mistakenly find the complementary strands.